Profiling
Summary
Name |
MPI |
OpenMP |
Cuda |
SIMD |
|---|---|---|---|---|
Advisor |
✓ |
✓ |
||
AMD µProf |
✓ |
✓ |
||
Linaro MAP |
✓ |
✓ |
✓ |
✓ |
Cube |
✓ |
✓ |
✓ |
✓ |
Darshan |
✓ |
✓ |
||
Gprof |
||||
cProfile |
||||
HPCToolkit |
✓ |
✓ |
||
Igprof |
||||
IPM |
✓ |
|||
Memonit |
✓ |
✓ |
||
SelFIe |
||||
Paraver |
✓ |
✓ |
✓ |
|
PAPI |
✓ |
|||
Perf |
✓ |
|||
ScoreP |
✓ |
✓ |
✓ |
|
Tau |
✓ |
✓ |
✓ |
|
Valgrind (cachegrind) |
✓ |
✓ |
||
Valgrind (callgrind) |
✓ |
✓ |
||
Valgrind (massif) |
✓ |
✓ |
||
Vampir |
✓ |
✓ |
✓ |
|
Vtune |
✓ |
✓ |
Name |
Comm |
I/O |
Call graph |
Hardware counters |
Memory usage |
Cache usage |
|---|---|---|---|---|---|---|
Advisor |
✓ |
|||||
AMD µProf |
✓ |
✓ |
✓ |
|||
Linaro MAP |
✓ |
✓ |
✓ |
|||
Cube |
✓ |
✓ |
✓ |
|||
Darshan |
✓ |
|||||
Gprof |
✓ |
|||||
cProfile |
✓ |
|||||
HPCToolkit |
✓ |
✓ |
✓ |
|||
Igprof |
✓ |
|||||
IPM |
✓ |
✓ |
||||
Memonit |
✓ |
|||||
SelFIe |
✓ |
✓ |
✓ |
✓ |
||
Paraver |
✓ |
✓ |
||||
PAPI |
✓ |
✓ |
✓ |
|||
Perf |
✓ |
✓ |
✓ |
✓ |
✓ |
|
ScoreP |
✓ |
✓ |
✓ |
✓ |
✓ |
|
Tau |
✓ |
✓ |
✓ |
✓ |
||
Valgrind (cachegrind) |
✓ |
|||||
Valgrind (callgrind) |
✓ |
|||||
Valgrind (massif) |
✓ |
|||||
Vampir |
✓ |
|||||
Vtune |
✓ |
✓ |
Name |
Collection |
GUI |
Sampling |
Tracing |
Instrumentation necessary |
|---|---|---|---|---|---|
Advisor |
✓ |
✓ |
✓ |
||
AMD µProf |
✓ |
✓ |
✓ |
✓ |
|
Linaro MAP |
✓ |
✓ |
✓ |
✓ |
|
Cube |
✓ |
✓ |
✓ |
✓ |
|
Darshan |
✓ |
✓ |
|||
Gprof |
✓ |
✓ |
✓ |
✓ |
|
cProfile |
✓ |
✓ |
✓ |
✓ |
|
HPCToolkit |
✓ |
✓ |
✓ |
✓ |
|
Igprof |
✓ |
✓ |
✓ |
||
IPM |
✓ |
✓ |
|||
Memonit |
✓ |
✓ |
✓ |
||
SelFIe |
✓ |
✓ |
|||
Paraver |
✓ |
✓ |
✓ |
||
PAPI |
✓ |
✓ |
✓ |
||
Perf |
✓ |
||||
ScoreP |
✓ |
✓ |
✓ |
✓ |
|
Tau |
✓ |
✓ |
✓ |
✓ |
|
Valgrind (cachegrind) |
✓ |
✓ |
|||
Valgrind (callgrind) |
✓ |
✓ |
|||
Valgrind (massif) |
✓ |
✓ |
|||
Vampir |
✓ |
✓ |
✓ |
||
Vtune |
✓ |
✓ |
✓ |
To display a list of all available profilers use the search option of the module command :
$ module search profiler
SelFIe
SelFIe (SElf and Light proFIling Engine) is a tool to lightly profile Linux commands without compiling. The profiling is done by a dynamic library which can be given to the LD_PRELOAD environment variable before the execution of the command. It doesn’t affect the behaviour of the command and users don’t see any changes at execution. At the end of the execution, it puts a line in system logs:
Note
SelFIe is a opensource software developed by CEA - Selfie on Github
selfie[26058]: { "utime": 0.00, "stime": 0.01, "maxmem": 0.00, "posixio_time": 0.00, "posixio_count": 7569, "USER": "user", "wtime": 0.01, "command": "/bin/hostname" }
To enable selfie:
$ module load feature/selfie/enable
To disable selfie:
$ module load feature/selfie/disable
To display the log in standard output:
$ module load feature/selfie/report
If your process takes less than 5 minutes, then load:
$ module load feature/selfie/short_job
SelFIe data can be written in a custom file by exporting the environment variable
SELFIE_OUTPUTFILE:
$ export SELFIE_OUTPUTFILE=selfie.data
$ ccc_mprun -p |default_CPU_partition| -Q test -T 300 ./a.out
$ cat selfie.data
selfie[2152192]: { "utime": 0.00, "stime": 0.00, "maxmem": 0.01, "hostname": "host1216", "posixio_time": 0.00, "posixio_count": 4, "mpi_time": 0.00, "mpi_count": 0, "mpiio_time": 0.00, "mpiio_count": 0, "USER": "username", "SLURM_JOBID": "3492220", "SLURM_STEPID": "0", "SLURM_PROCID": "0", "OMP_NUM_THREADS": "1", "timestamp": 1687937047, "wtime": 30.00, "command": "./a.out" }
Example of a short job in standard output:
$ module load feature/selfie/enable
$ module load feature/selfie/report
$ module load feature/selfie/short_job
$ ccc_mprun -p |default_CPU_partition| -Q test -T 300 ./a.out
selfie[2152192]: { "utime": 0.00, "stime": 0.00, "maxmem": 0.01, "hostname": "host1216", "posixio_time": 0.00, "posixio_count": 4, "mpi_time": 0.00, "mpi_count": 0, "mpiio_time": 0.00, "mpiio_count": 0, "USER": "username", "SLURM_JOBID": "3492220", "SLURM_STEPID": "0", "SLURM_PROCID": "0", "OMP_NUM_THREADS": "1", "timestamp": 1687937047, "wtime": 30.00, "command": "./a.out" }
You can format the JSON output via the jq command:
$ jq . <<< '{ "utime": 0.00, "stime": 0.00, "maxmem": 0.01, "hostname": "host1216", "posixio_time": 0.00, "posixio_count": 4, "mpi_time": 0.00, "mpi_count": 0, "mpiio_time": 0.00, "mpiio_count": 0, "USER": "username", "SLURM_JOBID": "3492220", "SLURM_STEPID": "0", "SLURM_PROCID": "0", "OMP_NUM_THREADS": "1", "timestamp": 1687937047, "wtime": 30.00, "command": "./a.out" }'
{
"utime": 0,
"stime": 0,
"maxmem": 0.01,
"hostname": "host1216",
"posixio_time": 0,
"posixio_count": 4,
"mpi_time": 0,
"mpi_count": 0,
"mpiio_time": 0,
"mpiio_count": 0,
"USER": "username",
"SLURM_JOBID": "3492220",
"SLURM_STEPID": "0",
"SLURM_PROCID": "0",
"OMP_NUM_THREADS": "1",
"timestamp": 1687937047,
"wtime": 30,
"command": "./a.out"
}
IPM
IPM is a light-weight profiling tool that profiles the mpi calls and memory usage in a parallel program. IPM cannot be used on a multi-threaded program.
To run a program with IPM profiling, just load the ipm module (no need to instrument or recompile anything) and run it with :
#!/bin/bash
#MSUB -r MyJob_Para # Job name
#MSUB -n 32 # Number of tasks to use
#MSUB -T 1800 # Elapsed time limit in seconds
#MSUB -o example_%I.o # Standard output. %I is the job id
#MSUB -e example_%I.e # Error output. %I is the job id
#MSUB -q <partition> # Partition name
#MSUB -A <project> # Project ID
set -x
cd ${BRIDGE_MSUB_PWD}
module load ipm
#The ipm module tells ccc_mprun to use IPM library
ccc_mprun ./prog.exe
It will generate a report at the end of the standard output of the job and an xml file. It is possible to generate a graphical and complete html page with the command:
$ ipm_parse -html XML_File
Example of IPM output
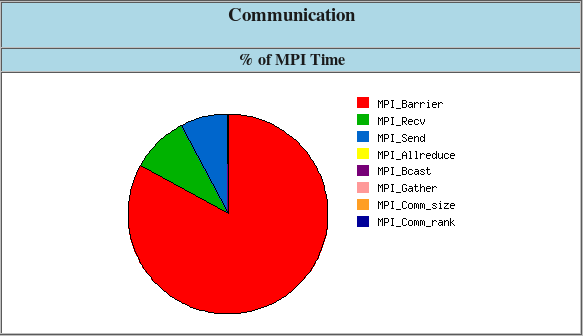
Example of IPM output
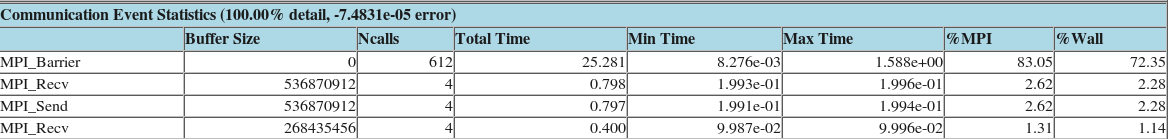
Linaro-forge MAP
Linaro-forge MAP is the profiler for parallel, multithreaded or single threaded C, C++ and F90 codes. MAP gives information on memory usage, MPI and OpenMP usage, percentage of vectorized SIMD instructions, etc.
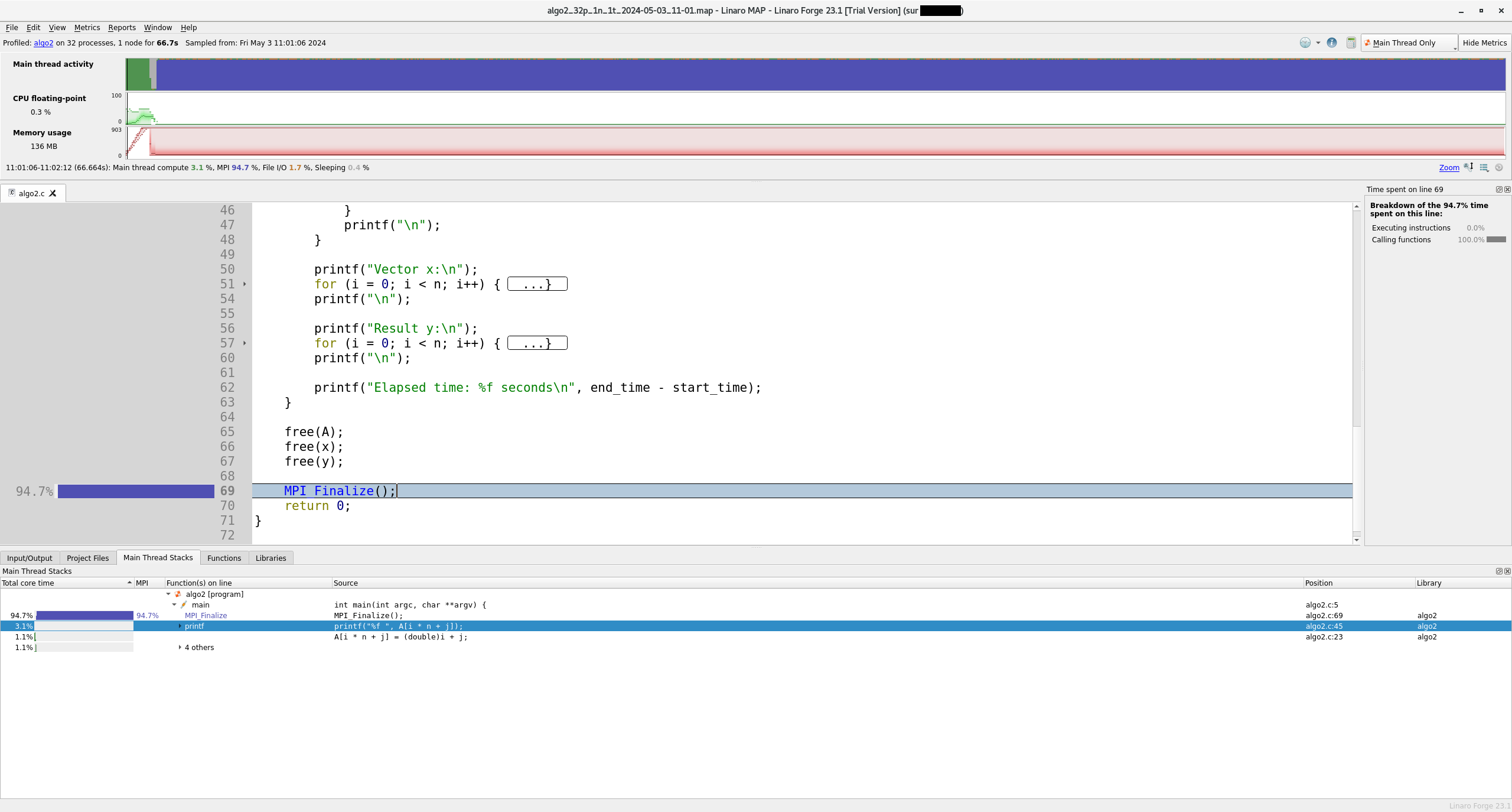
Example of MAP profile
The code just has to be compiled with -g for debugging information. No instrumentation is needed.
You can profile your code with map by loading the appropriate module:
$ module load linaro-forge
Then use the command map --profile. For parallel codes, edit your submission script and just replace ccc_mprun with map --profile.
Example of submission script:
#!/bin/bash
#MSUB -r MyJob_Para # Job name
#MSUB -q partition # Partition name
#MSUB -n 32 # Number of tasks to use
#MSUB -T 1800 # Elapsed time limit in seconds
#MSUB -o example_%I.o # Standard output. %I is the job id
#MSUB -e example_%I.e # Error output. %I is the job id
#MSUB -A <project> # Project ID
set -x
cd ${BRIDGE_MSUB_PWD}
module load linaro-forge
map --profile -n 32 ./a.out
Once the job has finished, a .map file should have been created. It can be opened from a remote desktop session or from an interactive session with the following command:
$ map <output_name>.map
Note
Linaro-forge MAP is a licenced product.
A full documentation is available in the installation path on the cluster. To open it:
$ evince ${MAP_ROOT}/doc/userguide-forge.pdf
Note
Allinea-forge has been renamed Arm-forge, which has then been renamed Linaro-forge.
Scalasca
Scalasca is a set of software which let you profile your parallel code by taking traces during the execution of the program. It is actually a wrapper that launches Score-P and Cube. This software is a kind of parallel gprof with more information. We present here an introduction of Scalasca. The generated output can then be opened with several analysis tools like Periscope, Cube, Vampir, or Tau.
Scalasca profiling requires 3 different steps:
Instrumenting the code with skin
Collecting profiling data with scan
Examine collected information with square
Code instrumentation with Scalasca
First step for profiling a code with is instrumentation. You must compile your code by adding the wrapper before the call to the compiler. You need to load the scalasca module beforehand :
$ module load scalasca
$ skin mpicc -g -c prog.c
$ skin mpicc -o prog.exe prog.o
or for Fortran :
$ module load scalasca
$ skin mpif90 -g -c prog.f90
$ skin mpif90 -o prog.exe prog.o
You can compile for OpenMP programs:
$ skin ifort -openmp -g -c prog.f90
$ skin ifort -openmp -o prog.exe prog.o
You can profile hybrid MPI-OpenMP programs:
$ skin mpif90 -openmp -g -O3 -c prog.f90
$ skin mpif90 -openmp -g -O3 -o prog.exe prog.o
Simple profiling with Scalasca
Once the code has been instrumented with Scalasca, run it with scan. By default, a simple summary profile is generated.
Here is a simple example of a submission script:
#!/bin/bash
#MSUB -r MyJob_Para # Job name
#MSUB -n 32 # Number of tasks to use
#MSUB -T 1800 # Elapsed time limit in seconds
#MSUB -o example_%I.o # Standard output. %I is the job id
#MSUB -e example_%I.e # Error output. %I is the job id
#MSUB -q <partition> # Partition name
#MSUB -A <project> # Project ID
set -x
cd ${BRIDGE_MSUB_PWD}
module load scalasca
export SCOREP_EXPERIMENT_DIRECTORY=scorep_profile.${BRIDGE_MSUB_JOBID}
scan ccc_mprun ./prog.exe
At the end of execution, the program generates a directory which contains the profiling files (the directory name is chosen with the SCOREP_EXPERIMENT_DIRECTORY environment variable):
$ tree scorep_profile.2871901
|- profile.cubex
`- scorep.cfg
The profile information can then be visualized with square:
$ module load scalasca
$ square scorep_profile.2871901
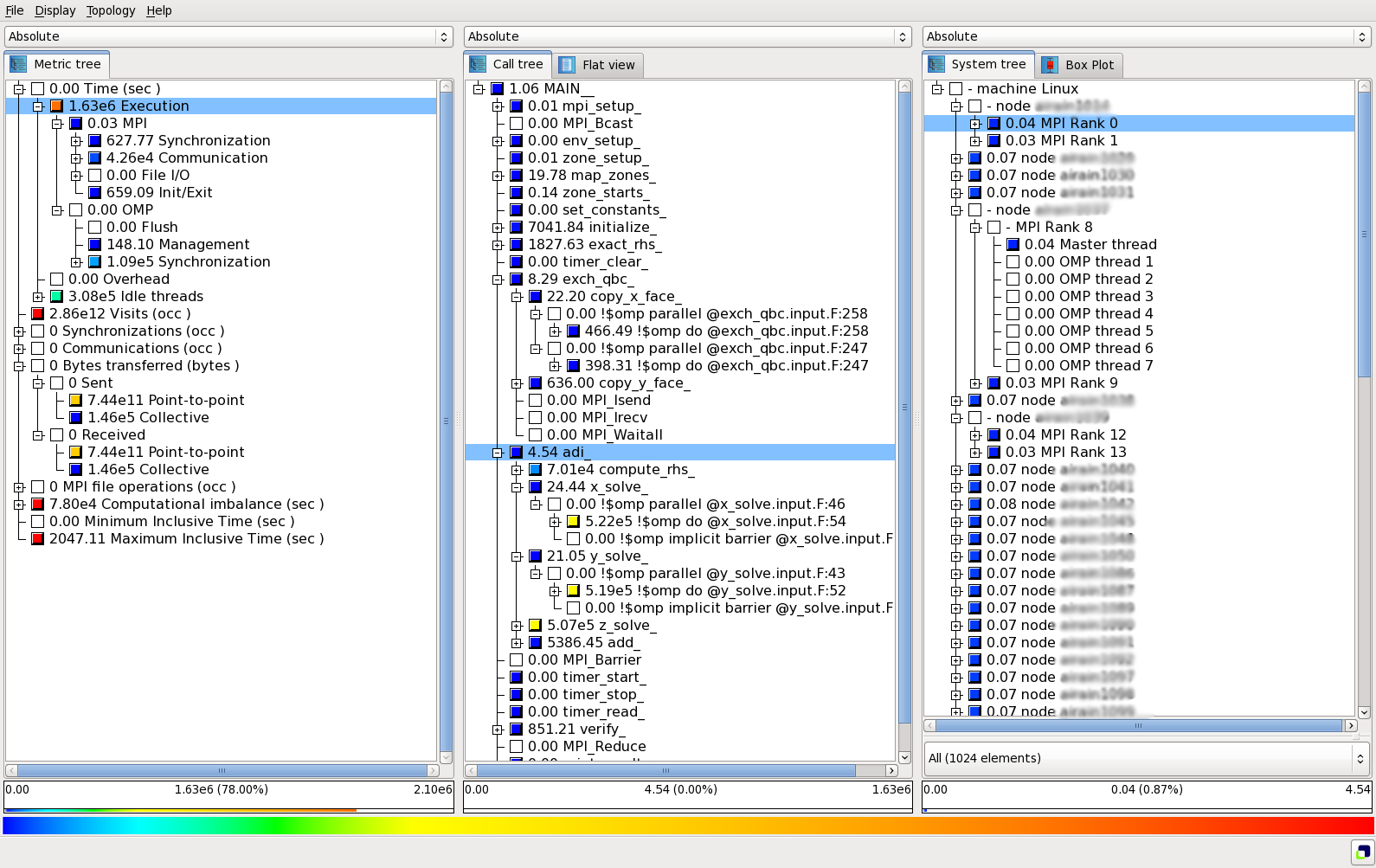
Cube interface
Scalasca with PAPI
Score-P can retrieve the hardware counter with PAPI. For example, if you want retrieve the number of floating point operations:
#!/bin/bash
#MSUB -r MyJob_Para # Job name
#MSUB -n 32 # Number of tasks to use
#MSUB -T 1800 # Elapsed time limit in seconds
#MSUB -o example_%I.o # Standard output. %I is the job id
#MSUB -e example_%I.e # Error output. %I is the job id
#MSUB -q <partition> # Partition name
#MSUB -A <project> # Project ID
set -x
cd ${BRIDGE_MSUB_PWD}
module load scalasca
export SCOREP_EXPERIMENT_DIRECTORY=scorep_profile.${BRIDGE_MSUB_JOBID}
export SCOREP_METRIC_PAPI=PAPI_FP_OPS
scan ccc_mprun ./prog.exe
Then the number of floating point operations will appear on the profile when you visualize it. The the syntax to use several papi counters is:
export SCOREP_METRIC_PAPI="PAPI_FP_OPS,PAPI_TOT_CYC"
Tracing application with Scalasca
To get a full trace there is no need to recompile the code. The same instrumentation is used for summary and trace profiling. To activate the trace collection, use the option -t of scan.
#!/bin/bash
#MSUB -r MyJob_Para # Job name
#MSUB -n 32 # Number of tasks to use
#MSUB -T 1800 # Elapsed time limit in seconds
#MSUB -o example_%I.o # Standard output. %I is the job id
#MSUB -e example_%I.e # Error output. %I is the job id
#MSUB -q <partition> # partition name
#MSUB -A <project> # Project ID
set -x
cd ${BRIDGE_MSUB_PWD}
module load scalasca
export SCOREP_EXPERIMENT_DIRECTORY=scorep_profile.${BRIDGE_MSUB_JOBID}
scan -t ccc_mprun ./prog.exe
In that case, a file traces.otf2 is created in the output directory with the summary. This profile trace can be opened with for example.
$ tree -L 1 scorep_profile.2727202
|-- profile.cubex
|-- scorep.cfg
|-- traces/
|-- traces.def
`-- traces.otf2
Warning
Generating a full trace may require a huge amount of memory.
Here is the best practice to follow:
First start with a simple Scalasca analysis (without
-t)Thanks to this summary, you can get an estimation of the size a full trace would take with the command:
$ square -s scorep_profile.2871901
Estimated aggregate size of event trace: 58GB
Estimated requirements for largest trace buffer (max_buf): 6GB
....
If the estimated aggregated size of event trace seems excessive (it can easily reach several TB), you will need to apply filtering before recording the trace.
For more information on filtering and profiling options, check out the full documentation provided in the installation path:
$ module load scalasca
$ evince ${SCALASCA_ROOT}/share/doc/scalasca/manual/UserGuide.pdf
Vampir
Vampir is a visualization software that can be used to analyse OTF traces. The traces should have been generated before by one of the available profiling software such as Score-P.
Usage
To open a Score-P trace with vampir, just launch the graphical interface with the corresponding OTF file.
$ module load vampir
$ vampir scorep_profile.2871915/traces.otf2
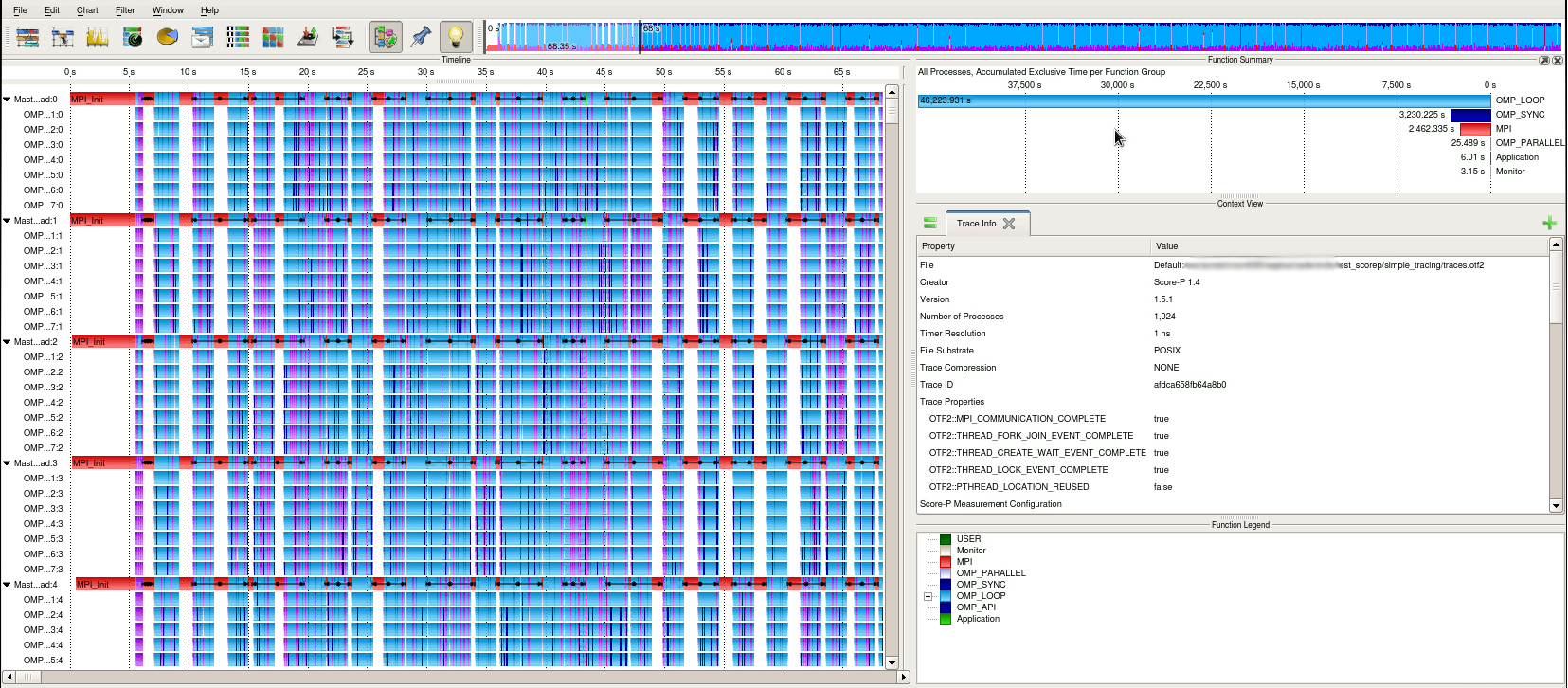
Vampir window
It is not recommended to launch on the login nodes. An interactive session on compute nodes may be necessary. Also, the graphical interface may be slow. Using the Remote Desktop System service can help with that.
See the manual for full details and features of the vampir tool.
Vampirserver
Traces generated by Score-P can be very large and can be very slow if you want to visualize these traces. Vampir provides vampirserver: it is a parallel program which uses CPU computing to accelerate Vampir visualization. Firstly, you have to submit a job which will launch on Irene nodes:
$ cat vampirserver.sh
#!/bin/bash
#MSUB -r vampirserver # Job name
#MSUB -n 32 # Number of tasks to use
#MSUB -T 1800 # Elapsed time limit in seconds
#MSUB -o vampirserver_%I.o # Standard output. %I is the job id
#MSUB -e vampirserver_%I.e # Error output. %I is the job id
#MSUB -q partition # Partition
#MSUB -A <project> # Project ID
module load vampirserver
vampirserver start -n $((BRIDGE_MSUB_NPROC-1))
sleep 1700
$ ccc_msub vampirserver.sh
When the job is running, you will obtain this output:
$ ccc_mpp
USER ACCOUNT BATCHID NCPU QUEUE PRIORITY STATE RLIM RUN/START SUSP OLD NAME NODES
toto genXXX 234481 32 large 210332 RUN 30.0m 1.3m - 1.3m vampirserver node1352
$ ccc_mpeek 234481
Found license file: /usr/local/vampir-7.5/bin/lic.dat
Running 31 analysis processes... (abort with Ctrl-C or vngd-shutdown)
Server listens on: node1352:30000
And a Vampir window should open.
Note
The vampirserver command runs in the background. So, without the call to Vampir, the job would be terminated immediately.
In our example, the Vampirserver master node is on node1352. The port to connect is 30000. Now you can use the graphical interface: start Vampir on a remote desktop service. Instead of clicking on “Open”, you will click on “Remote Open”:
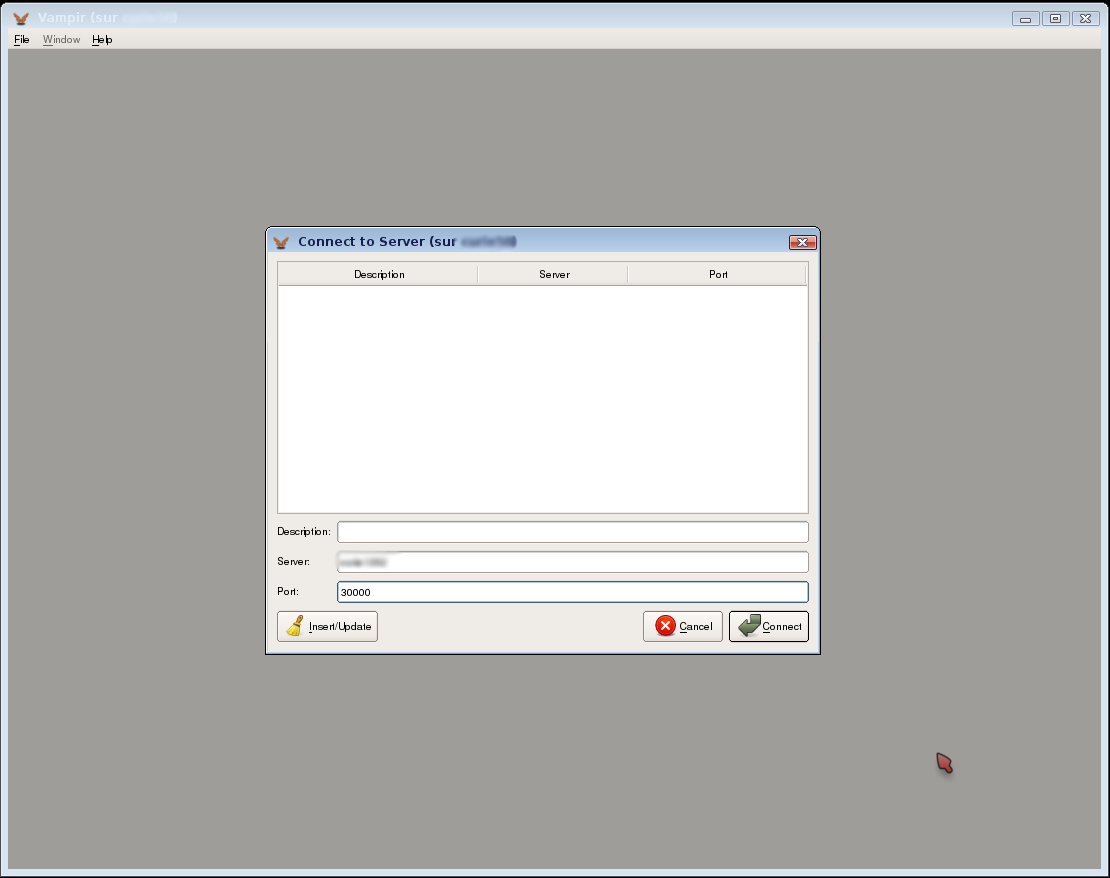
Connecting to Vampirserver
Fill the server and the port, for instance Server : node1352 and Port : 30000 in the previous case. You will be connected to vampirserver. Then you can open an OTF file and visualize it.
Note
You can ask any number of processors you want: it will be faster if your profiling files are big. But be careful, it consumes your computing times.
Don’t forget to delete the Vampirserver job after your analyse.
Score-P
Score-P is a software system that provides measurement infrastructure for profiling, event trace recording, and analysis of HPC applications. Its goal is to simplify the analysis of the behavior of HPC software and to allow developers to find out where and why performance problems arise, where bottlenecks may be expected and where their codes offer room for further improvements. Score-P supports MPI, OpenMP, Pthreads, and CUDA, OpenCL and OpenACC and Fortran, C and C++ languages. It can take traces (OTF2) to be read by Vampir or Scalasca, and call-path profiles to be read by CUBE or TAU.
Instrumentation
Score-P is available through the module command:
$ module load scorep
You need to compile your code with scorep. Compile and link with:
scorep mpicc for C code source files,
scorep mpicxx for C++ code source files, and
scorep mpif90 for Fortran code source files,
Usage
The command to run scorep is:
$ ccc_mprun -n $NPROCS ./my_app
When running the instrumented exectuable, the measurement system will create a directory called scorep-YYYYMMDD_HHMM_XXXXXXX, containing the date and time, and where XXXXXXXX is an additional identification number. The environment variables SCOREP_ENABLE_TRACING and SCOREP_ENABLE_PROFILING control whether event tracing or profiles are stored in this directory. By default, profiling is enabled and tracing is disabled.
After the completion of the execution, the requested data is available in the dedicated location, and can be visualized and analized by CUBE, TAU, or Vampir, for example.
For more information, see Score-P User Guide.
Darshan
Darshan is a scalable HPC I/O characterization tool. It is designed to profile I/O behavior with minimum overhead.
To run a program with Darshan profiling, there is no need to instrument or recompile the code.
Load the darshan module : it tells ccc_mprun to wrap the I/O calls with the Darshan library.
Specify where you want the darshan trace to be created by exporting the variable
DARSHAN_LOG_PATH.The path must point to one directory of your home
$CCCHOME.
Here is an example of a submission script to enable darshan profiling:
#!/bin/bash
#MSUB -r MyJob_Para # Job name
#MSUB -n 32 # Number of tasks to use
#MSUB -T 1800 # Elapsed time limit in seconds
#MSUB -o example_%I.o # Standard output. %I is the job id
#MSUB -e example_%I.e # Error output. %I is the job id
#MSUB -q <partition> # Partition
#MSUB -A <project> # Project ID
set -x
cd ${BRIDGE_MSUB_PWD}
module load darshan
export DARSHAN_LOG_PATH=$CCCHOME
#The darshan module tells ccc_mprun to use the Darshan library.
ccc_mprun ./prog.exe
This will generate a trace in the specified directory. Here is the format of the output file: <USERNAME>_<BINARY_NAME>_<JOB_ID>_<DATE>_<UNIQUE_ID>_<TIMING>.darshan.gz.
Some scripts are available to post-process the output. For instance, darshan-parser, darshan-job-summary.pl and darshan-summary-per-file.sh.
darshan-job-summary.pl will generate a graphical summary of the I/O activity for a job.
$ darshan-job-summary.pl *.darshan.gz
darshan-summary-per-file.sh is similar except that it produces a separate pdf summary for every file accessed by the application. The summaries will be written in the directory specified as argument.
$ darshan-summary-per-file.sh *.darshan.gz output_dir
darshan-parser gives a full, human readable dump of all information contained in a log file.
$ darshan-parser *.darshan.gz > example_output.txt
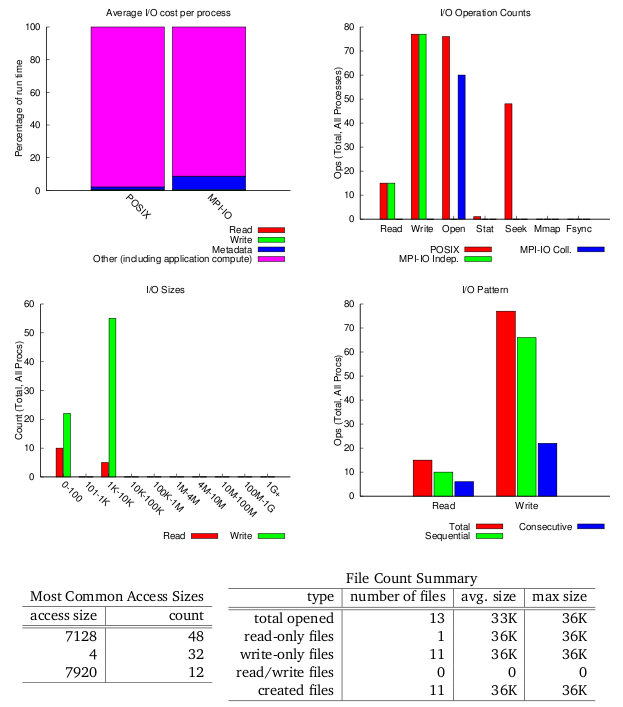
Example of Darshan output
PAPI
PAPI is an API which allows you to retrieve hardware counters from the CPU. Here an example in Fortran to get the number of floating point operations of a matrix DAXPY:
program main
implicit none
include 'f90papi.h'
!
integer, parameter :: size = 1000
integer, parameter :: ntimes = 10
double precision, dimension(size,size) :: A,B,C
integer :: i,j,n
! Variable PAPI
integer, parameter :: max_event = 1
integer, dimension(max_event) :: event
integer :: num_events, retval
integer(kind=8), dimension(max_event) :: values
! Init PAPI
call PAPIf_num_counters( num_events )
print *, 'Number of hardware counters supported: ', num_events
call PAPIf_query_event(PAPI_FP_INS, retval)
if (retval .NE. PAPI_OK) then
event(1) = PAPI_TOT_INS
else
! Total floating point operations
event(1) = PAPI_FP_INS
end if
! Init Matrix
do i=1,size
do j=1,size
C(i,j) = real(i+j,8)
B(i,j) = -i+0.1*j
end do
end do
! Set up counters
num_events = 1
call PAPIf_start_counters( event, num_events, retval)
! Clear the counter values
call PAPIf_read_counters(values, num_events,retval)
! DAXPY
do n=1,ntimes
do i=1,size
do j=1,size
A(i,j) = 2.0*B(i,j) + C(i,j)
end do
end do
end do
! Stop the counters and put the results in the array values
call PAPIf_stop_counters(values,num_events,retval)
! Print results
if (event(1) .EQ. PAPI_TOT_INS) then
print *, 'TOT Instructions: ',values(1)
else
print *, 'FP Instructions: ',values(1)
end if
end program main
To compile, you have to load the PAPI module:
bash-4.00 $ module load papi/4.2.1
bash-4.00 $ ifort ${PAPI_CFLAGS} papi.f90 ${PAPI_LDFLAGS}
bash-4.00 $ ./a.out
Number of hardware counters supported: 7
FP Instructions: 10046163
To get the available hardware counters, you can type papi_avail command.
This library can retrieve the MFLOPS of a certain region of your code:
program main
implicit none
include 'f90papi.h'
!
integer, parameter :: size = 1000
integer, parameter :: ntimes = 100
double precision, dimension(size,size) :: A,B,C
integer :: i,j,n
! Variable PAPI
integer :: retval
real(kind=4) :: proc_time, mflops, real_time
integer(kind=8) :: flpins
! Init PAPI
retval = PAPI_VER_CURRENT
call PAPIf_library_init(retval)
if ( retval.NE.PAPI_VER_CURRENT) then
print*, 'PAPI_library_init', retval
end if
call PAPIf_query_event(PAPI_FP_INS, retval)
! Init Matrix
do i=1,size
do j=1,size
C(i,j) = real(i+j,8)
B(i,j) = -i+0.1*j
end do
end do
! Setup Counter
call PAPIf_flips( real_time, proc_time, flpins, mflops, retval )
! DAXPY
do n=1,ntimes
do i=1,size
do j=1,size
A(i,j) = 2.0*B(i,j) + C(i,j)
end do
end do
end do
! Collect the data into the Variables passed in
call PAPIf_flips( real_time, proc_time, flpins, mflops, retval)
! Print results
print *, 'Real_time: ', real_time
print *, ' Proc_time: ', proc_time
print *, ' Total flpins: ', flpins
print *, ' MFLOPS: ', mflops
!
end program main
and the output:
bash-4.00 $ module load papi/4.2.1
bash-4.00 $ ifort ${PAPI_CFLAGS} papi_flops.f90 ${PAPI_LDFLAGS}
bash-4.00 $ ./a.out
Real_time: 6.1250001E-02
Proc_time: 5.1447589E-02
Total flpins: 100056592
MFLOPS: 1944.826
If you want more precisions, you can contact us or visit PAPI website.
Memonit
Memonit is a memory profiling tool for HPC jobs.
It traces memory usage and can be used to plot allocated memory for each process or node across the execution of an MPI program.
To use the memonit tool:
Load the memonit module in your submission script:
module load memonit
If needed, change snapshot frequency:
export MEMONIT_DELAY=0:100 # snapshot every 100us, by default every 2s
Insert a call to the memonit_collect wrapper when launching your computation:
ccc_mprun memonit_collect ./my_job
Process the collected traces for post-treatment:
After your job is complete, records have been written to a collection directory.
Use memonit -a <prefix> to consolidate and aggregate these records using the “prefix” name of the directory (if the directory name is my_job.<JobId>.d, use my_job.<JobId> as the prefix name), and memonit -g or memonit_gui to open the results with the graphic interface (GUI requires X forwarding).
Both steps can be combined in a single command, such as memonit -a -g <prefix>.
Example of lines that you can insert into your submission script:
# Load dependencies
module load python
# Aggregate traces into my_job.db
memonit_aggregate -f my_job.metadata.json
# Analyse by mpi rank
memonit_gui -f my_job.db
# Or by node
memonit_gui -f my_job_bynode.db
Valgrind
Valgrind is an instrumentation framework for dynamic analysis tools. It comes with a set of tools for profiling and debugging codes.
To run a program with valgrind, there is no need to instrument, recompile, or otherwise modify the code.
Note
A tool in the Valgrind distribution can be invoked with the --tool option:
module load valgrind
valgrind --tool=<toolname> #default is memcheck
Callgrind
Callgrind is a profiling tool that records the call history among functions in a program’s run as a call-graph. By default, the collected data consists of the number of instructions executed, their relationship to source lines, the caller/callee relationship between functions, and the numbers of such calls.
To start a profile run for a program, execute:
module load valgrind
valgrind --tool=callgrind [callgrind options] your-program [program options]
While the simulation is running, you can observe the execution with:
callgrind_control -b
This will print out the current backtrace. To annotate the backtrace with event counts, run:
callgrind_control -e -b
After program termination, a profile data file named callgrind.out.<pid> is generated, where pid is the process ID of the program being profiled. The data file contains information about the calls made in the program among the functions executed, together with Instruction Read (Ir) event counts.
To generate a function-by-function summary from the profile data file, use:
callgrind_annotate [options] callgrind.out.<pid>
Cachegrind
Cachegrind simulates how your program interacts with a machine’s cache hierarchy and (optionally) branch predictor. To run cachegrind on a program prog, you must specify --tool=cachegrind on the valgrind command line:
module load valgrind
valgrind --tool=cachegrind prog
Branch prediction statistics are not collected by default. To do so, add the option --branch-sim=yes.
valgrind -tool=cachegrind --branch-sim=yes prog
One output file will be created for each process launched with cachegrind. To analyse the output, use the command cg_annotate:
$ cg_annotate <output_file>
cg_annotate can show the source codes annotated with the sampled values. Therefore, either use the option --auto=yes to apply to all the available source files or specify one file by passing it as an argument.
$ cg_annotate <output_file> sourcecode.c
For more information on Cachegrind, check out the official Cachegrind User Manual.
Massif
Massif measures how much heap memory a program uses. This includes both the useful space, and the extra bytes allocated for book-keeping and alignment purposes. Massif can optionally measure the stack memory.
As for the other Valgrind tools, the program should be compiled with debugging info (the -g option). To run massif on a program prog, the valgrind command line is:
module load valgrind
valgrind --tool=massif prog
The Massif option --pages-as-heap=yes allows to measure all the memory used by the program.
By default, the output file is called massif.out.<pid> (pid is the process ID), although this file name can be changed with the --massif-out-file option. To present the heap profiling information about the program in a readable way, run ms_print:
$ ms_print massif.out.<pid>
TAU (Tuning and Analysis Utilities)
TAU Performance System is a portable profiling and tracing toolkit for performance analysis of parallel programs written in Fortran, C, C++, UPC, Java, Python.
TAU (Tuning and Analysis Utilities) is capable of gathering performance information through instrumentation of functions, methods, basic blocks, and statements as well as event-based sampling. The instrumentation can be inserted in the source code using an automatic instrumentor tool based on the Program Database Toolkit (PDT), dynamically using DyninstAPI, at runtime in the Java Virtual Machine, or manually using the instrumentation API.
TAU’s profile visualization tool, paraprof, provides graphical displays of all the performance analysis results, in aggregate and single node/context/thread forms. The user can quickly identify sources of performance bottlenecks in the application using the graphical interface. In addition, TAU can generate event traces that can be displayed with the Vampir, Paraver or JumpShot trace visualization tools.
Instrumentation
TAU is available through the module command:
$ module load tau
Specify programming model by setting TAU_MAKEFILE to one of $TAU_MAKEFILEDIR/Makefile.tau-*:
Makefile.tau-icpc-papi-mpi-cupti-pdt-openmp
Compile and link with:
tau_cc.sh for C code source files,
tau_cxx.sh for C++ code source files, and
tau_f90.sh for Fortran code source files,
Usage
The command to run TAU is:
$ ccc_mprun -n $NPROCS tau_exec ./a.out
Examine results with paraprof/pprof
$ pprof [directory_path]
or
$ paraprof
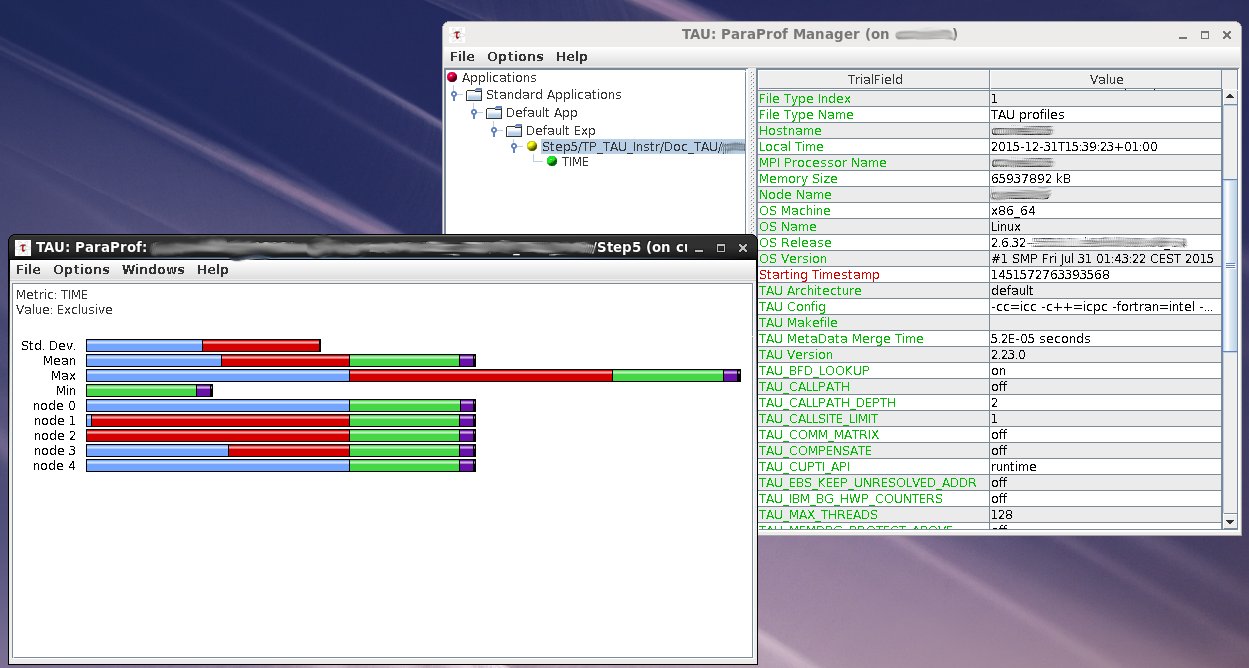
Example of ParaProf window
Note
It’s recommended to use the Remote Desktop session to use the graphical tools provided by tau (e.g. paraprof).
Environment variables control measurement mode TAU_PROFILE, TAU_TRACE, TAU_CALLPATH are available to tune the profiling settings.
For more information, see TAU User Guide.
Perf
Perf is a portable tool included in Linux kernel, it doesn’t need to be loaded, doesn’t need any driver and also works on all Linux platforms.
It is a performance analysis tool which displays performance counters such as the number of cache loads misses or branch loads misses.
Perf cannot be fully launched on login nodes, you will need to execute it on compute node(s).
In order to profile several processes with perf, you will need to differentiate the outputs by processe. Here is a simple wrapper to do it :
$ cat wrapper_perf.sh
#!/bin/bash
perf record -o out.${SLURM_PROCID}.data $@
To run a command and gather performance counter statistics use perf stat. Here is a simple job example:
#!/bin/bash
#MSUB -n 4
#MSUB -c 12
#MSUB -T 400
#MSUB -q |Partition|
#MSUB -A <project>
#MSUB -x
#MSUB -E '--enable_perf'
export OMP_NUM_THREADS=${BRIDGE_MSUB_NCORE}
ccc_mprun wrapper_perf stat ./exe
Informations will be stored in
out.<jobID>.data:
$ cat out.0.data
# started on Wed Oct 11 14:01:14 2017
Performance counter stats for './exe':
50342.076495 task-clock:u (msec) # 1.000 CPUs utilized
98,323 page-faults:u # 0.002 M/sec
155,516,791,925 cycles:u # 3.089 GHz
197,715,764,466 instructions:u # 1.27 insn per cycle
50.348439743 seconds time elapsed
Now for more specifics counters:
list of all performance counters with the command perf list
Use the
-eoption to specify the wanted performance counters. For example, to get how much cache access misses within all cache access:
$ ccc_mprun perf stat -o perf.log -e cache-references,cache-misses ./exe
$ cat perf.log
# started on Wed Oct 11 14:02:52 2017
Performance counter stats for './exe':
8,654,163,728 cache-references:u
875,346,349 cache-misses:u # 10.115 % of all cache refs
52.710267128 seconds time elapsed
Perf can also record few counters from an executable and create a report:
Use a job to create a report with
$ ccc_mprun -vvv perf record -o data.perf ./exe
...
[ perf record: Woken up 53 times to write data ]
[ perf record: Captured and wrote 13.149 MB data.perf (344150 samples) ]
...
Read the report from the login node
$ perf report -i data.perf
Samples: 344K of event 'cycles:u', Event count (approx.): 245940413046
Overhead Command Shared Object Symbol
99.94% exe exe [.] mydgemm_
0.02% exe [kernel.vmlinux] [k] apic_timer_interrupt
0.02% exe [kernel.vmlinux] [k] page_fault
0.00% exe exe [.] MAIN__
...
You can also using call graphs with
$ ccc_mprun -vvv perf record --call-graph fp -o data.perf ./exe
$ perf report -i data.perf
Samples: 5K of event 'cycles:u', Event count (approx.): 2184801676
Children Self Command Shared Object Symbol
+ 66.72% 0.00% exe libc-2.17.so [.] __libc_start_main
+ 61.03% 0.03% exe libiomp5.so [.] __kmpc_fork_call
- 60.96% 0.05% exe libiomp5.so [.] __kmp_fork_call
60.90% __kmp_fork_call
- __kmp_invoke_microtask
56.06% nextGen
3.33% main
1.35% __intel_avx_rep_memcpy
+ 60.90% 0.03% exe libiomp5.so [.] __kmp_invoke_microtask
+ 56.06% 56.06% exe exe [.] nextGen
+ 8.98% 5.86% exe exe [.] main
...
Extra-P
Extra-P is an automatic performance-modeling tool that can be used to analyse several SCOREP_EXPERIMENT_DIRECTORY generated with Score-P. The primary goal of this tool is identify scalability bugs but due to his multiple graphics outputs, it’s also useful to make reports.
Usage
To analyse the scalability of an algorithm you need to generate several SCOREP_EXPERIMENT_DIRECTORY, for example you can launch this submission script:
#!/bin/bash
#MSUB -r npb_btmz_scorep
#MSUB -o npb_btmz_scorep_%I.o
#MSUB -e npb_btmz_scorep_%I.e
#MSUB -Q test
#MSUB -T 1800 # max walltime in seconds
#MSUB -q haswell
#MSUB -A <project>
cd $BRIDGE_MSUB_PWD
# benchmark configuration
export OMP_NUM_THREADS=$BRIDGE_MSUB_NCORE
# Score-P configuration
export SCOREP_EXPERIMENT_DIRECTORY=scorep_profile.p$p.r$r
PROCS=$BRIDGE_MSUB_NPROC
EXE=./exe
ccc_mprun -n $PROCS $EXE
from a bash script (4 runs on each scripts with 8, 16, 32 and 64 cores):
p=4 n=2 c=2 r=1 ccc_msub -n 2 -c 2 submit_global.msub
p=4 n=2 c=2 r=2 ccc_msub -n 2 -c 2 submit_global.msub
p=4 n=2 c=2 r=3 ccc_msub -n 2 -c 2 submit_global.msub
p=4 n=2 c=2 r=4 ccc_msub -n 2 -c 2 submit_global.msub
p=8 n=2 c=4 r=1 ccc_msub -n 2 -c 4 submit_global.msub
[...]
p=32 n=8 c=4 r=4 ccc_msub -n 8 -c 4 submit_global.msub
p=64 n=8 c=8 r=1 ccc_msub -n 8 -c 8 submit_global.msub
p=64 n=8 c=8 r=2 ccc_msub -n 8 -c 8 submit_global.msub
p=64 n=8 c=8 r=3 ccc_msub -n 8 -c 8 submit_global.msub
p=64 n=8 c=8 r=4 ccc_msub -n 8 -c 8 submit_global.msub
Once these folders are generated load Extra-P:
$ module load extrap
launch it and open the parent folder. The software detects automatically your files:
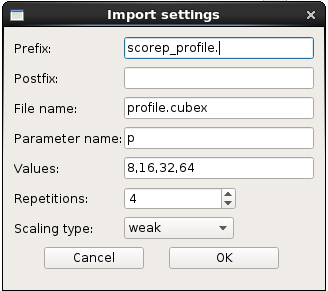
On Metric section choose “time” and you will get the time of each commands. You can also right clic on the graph and choose “Show all data points”, you will see the time of all your runs:
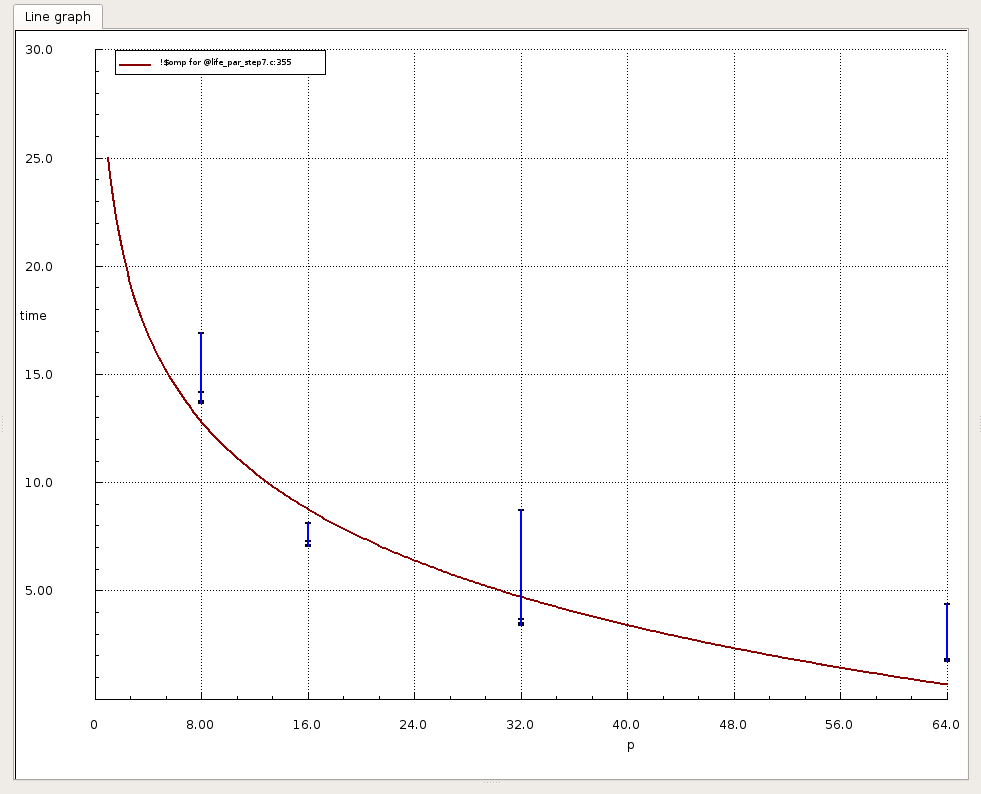
Note
These graphics only give the time of your application, not any speedup.
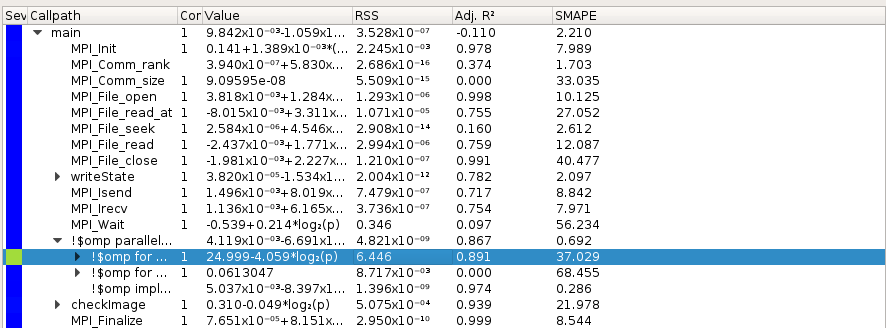
Extra-P also list all MPI and OpenMP requests, you can select what you want and check scalability:
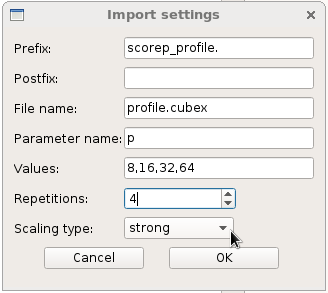
At least, you can choose to use strong scaling and see the efficiency of an algorithm:
AMD µProf
AMD µprof (“MICRO-prof”) is a performance analysis tool-suite for x86 based applications running on Linux. It provides performance metrics for AMD “Zen”-based processors and AMD Instinct MI Series accelerators. AMD µProf enables the developer to better understand the performance bottlenecks, optimization scope, and evaluate improvements. It offers the following functionalities:
Performance Analysis (CPU Profile): To identify runtime performance bottlenecks of the application.
System Analysis: To monitor system performance metrics, such as IPC and memory bandwidth.
Live Power Profile: To monitor thermal and power characteristics of the system.
$ module load uprof/3.4
Be careful, AMD µProf works in the /tmp directory. Therefore, you should copy your input files into it and copy your output files back to your base folder.
For CLI usage, here below is an example of submission script with several features of µprof you can test:
Example of submission script:
$ #!/bin/bash
#MSUB -r MyJob_Para # Job name
#MSUB -q partition # Partition name
#MSUB -n 32 # Number of tasks to use
#MSUB -T 1800 # Elapsed time limit in seconds
#MSUB -o example_%I.o # Standard output. %I is the job id
#MSUB -e example_%I.e # Error output. %I is the job id
#MSUB -A <project> # Project ID
set -x
cd ${BRIDGE_MSUB_PWD}
module load uprof
# Output directory
mkdir uprof_${BRIDGE_MSUB_JOBID}
# Copy all your executables in /tmp because AMDuProf works on it
cp -R ${BRIDGE_MSUB_PWD}/Examples/AMDTClassicMatMul /tmp/
# Profiling Time-Based Sampling
ccc_mprun AMDuProfCLI collect --config tbp -o /tmp/cpuprof-tbp -a Examples/AMDTClassicMatMul/bin/AMDTClassicMatMul-bin
wait
# Generate Reports
ccc_mprun AMDuProfCLI report -i /tmp/cpuprof-tbp.caperf -o /tmp/tbp_report.csv
wait
# Before being deleted, copy all your output CSV files from /tmp into your base directory
cp -R /tmp/tbp_report.csv/cpuprof-tbp ${BRIDGE_MSUB_PWD}/uprof_${BRIDGE_MSUB_JOBID}/
For interactive profiling, you can use AMDuProf in your submission script instead of AMDuProfCLI.
$ ccc_mprun AMDuProf Examples/AMDTClassicMatMul/bin/AMDTClassicMatMul-bin
First, connect to the interactive AMDuProf session. Then, select the instance you want to profile and your application target.
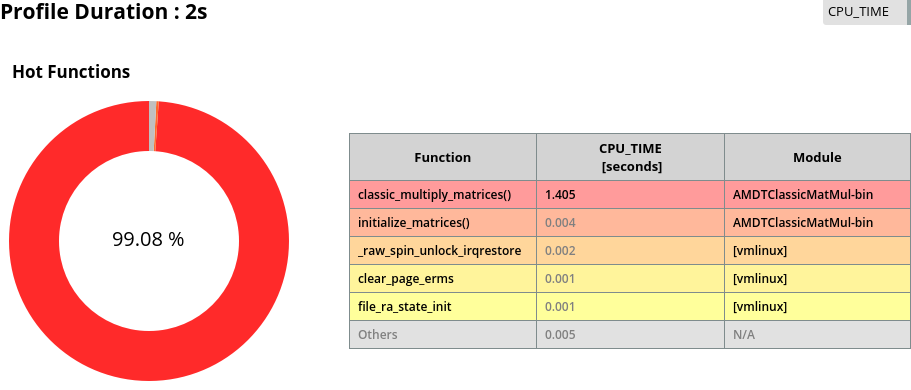
Example of AMD µProf interface
After a short time of profiling, you will see a detailed analysis of all of your execution instances.
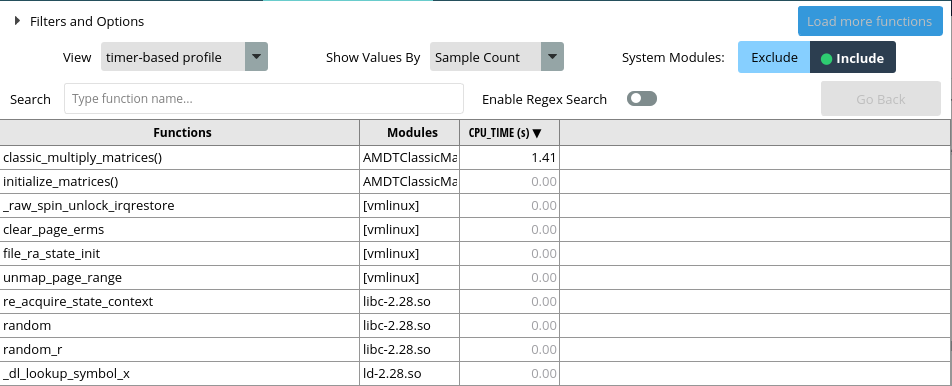
Some features of AMD µProf
AMDCpuTopology: Command-line tool to get the CPU topology of AMD Processors.
$ AMDCpuTopology
-----------------------------------------------
Package Numa CCD CCX Core Thread
-----------------------------------------------
0 0 0 0 0 0
0 0 0 0 0 64
... ... ... ... ... ....
Note
Please run all the following AMD µProf commands with ccc_mprun in your submission script to avoid root errors.
AMDuProfCLI: Command-line tool for µprof with profiling on AMD CPUs and GPUs.
AMDuProfCLI collect --config ...: Run a command and collect the performance profile data in .caperf file.
AMDuProfCLI timechart --event ... --interval ... --duration ...: Tool to visualize total system behavior during a workload like power, thermal and frequency.
AMDuProfCLI report -s event=... -i mon_programme.caperf: Process .caperf profile-data file created by AMDuProfCLI collect and generates the profile report.
AMDuProfCLI info ...: Run a command and display generic information about system, CPU etc.
Run AMDuProfCLI <COMMAND> -h for more information on a specific command.
AMDuProf: Graphical User Interface for intuitive visualization of performance metrics.
Timelines: Hardware-accelerated views; thread callstack tracing.
GPU Metrics: SMI data (power, temp, VRAM); HIP/HSA summary tables.
Source View: Reorder by execution/line number for optimized code analysis.
Gprof
Gprof produces an execution profile of C/C++ and Fortran programs. The effect of called routines is incorporated in the profile of each caller. The profile data is taken from the call graph profile file (gmon.out default) which is created by programs that are compiled with the -pg option. Gprof reads the given object file (the default is “a.out”) and establishes the relation between its symbol table and the call graph profile from gmon.out.
To load gprof:
$ module load gprof
To compile with
-pgoption
$ icc -pg hello.c
If your application is MPI, set environment variable to rename gmon files and enable one file per process
$ export GMON_OUT_PREFIX='gmon.out-'`/bin/uname -n`
Files generated will be named gmon.out-<hostname>.<pid> (ie: gmon.out-node1192.56893).
To generate call graph profile
gmon.out
$ ./a.out
To display flat profile and call graph
$ gprof a.out gmon.out
To display only the flat profile
$ gprof -p -b ./a.out gmon.out
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total
time seconds seconds calls s/call s/call name
67.72 10.24 10.24 1 10.24 10.24 bar
33.06 15.24 5.00 main
0.00 15.24 0.00 1 0.00 10.24 foo
To display the flat profile of one specific routine
$ gprof -p<routine> -b ./a.out gmon.out
To display only the call graph
$ gprof -q -b ./a.out gmon.out
Call graph (explanation follows)
index % time self children called name
<spontaneous>
[1] 100.0 5.00 10.24 main [1]
0.00 10.24 1/1 foo [3]
-----------------------------------------------
10.24 0.00 1/1 foo [3]
[2] 67.2 10.24 0.00 1 bar [2]
-----------------------------------------------
0.00 10.24 1/1 main [1]
[3] 67.2 0.00 10.24 1 foo [3]
10.24 0.00 1/1 bar [2]
-----------------------------------------------
To display the call graph of one specific routine
$ gprof -q<routine> -b ./a.out gmon.out
To generate a graph from data, see gprof2dot and gprof
Gprof2dot
Gprof2dot is an utility which converts profile data into a dot graph. It is compatible with many profilers.
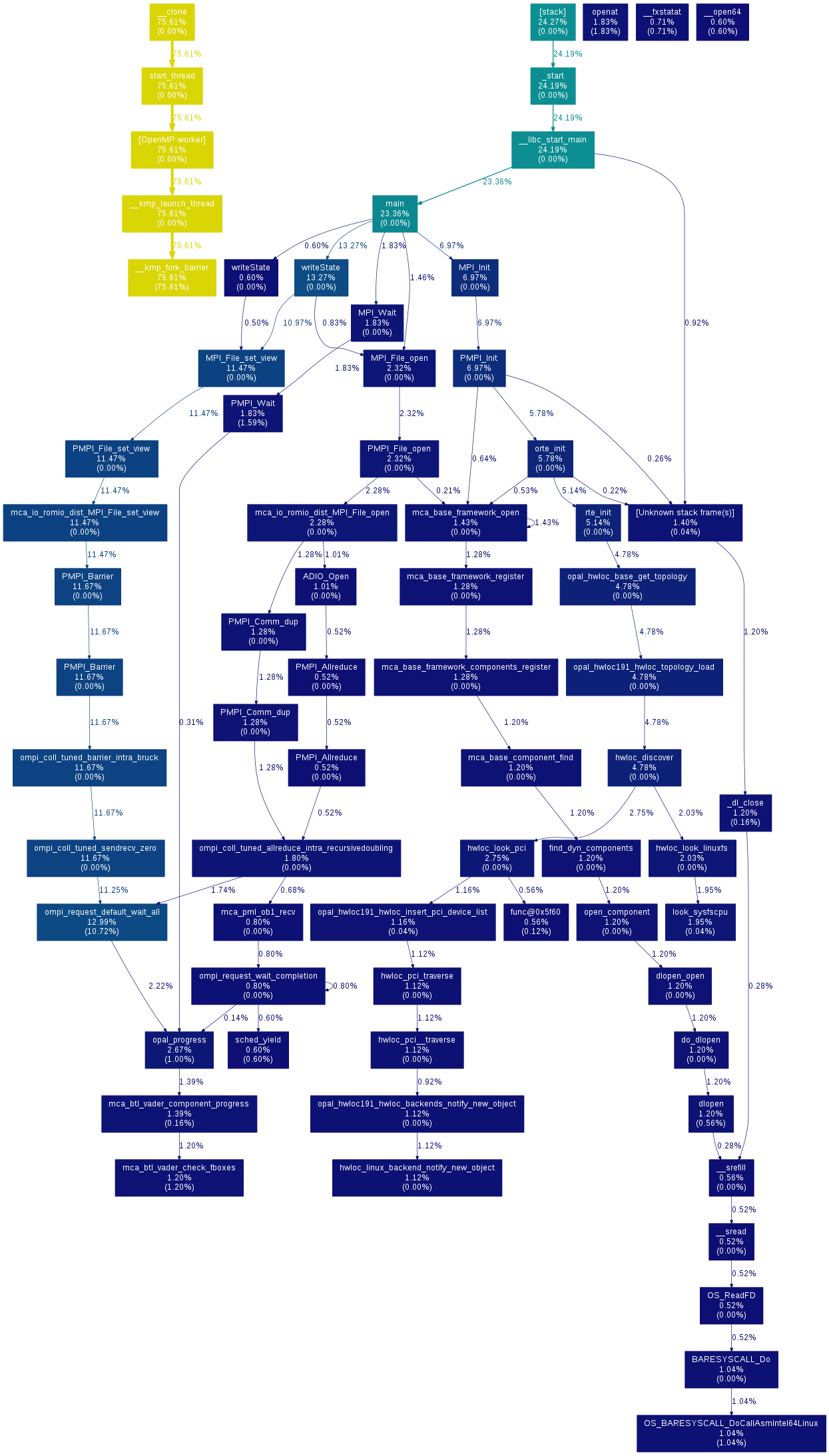
Use with gprof
Load modules gprof AND gprof2dot
$ module load gprof gprof2dot
Be sure to compile your application with
-pgoption
$ icc -pg hello.c
Generate the call graph profile
gmon.out(Run the application one time)
$ ./a.out
Generate the dot graph in a PNG image
$ gprof ./a.out gmon.out | gprof2dot.py | dot -Tpng -o a.png
Use with VTune in command line
Load modules VTune AND gprof2dot
$ module load vtune gprof2dot
Use VTune to collect data
$ ccc_mprun amplxe-cl -collect hotspots -result-dir output -- ./exe
Transform data in a gprof-like file
$ amplxe-cl -report gprof-cc -result-dir output -format text -report-output output.txt
Generate the dot graph
$ gprof2dot.py -f axe output.txt | dot -Tpng -o output.png
For more information about gprof2dot, see GitHub gprof2dot.
cProfile: Python profiler
cProfile is a profiler included with Python. It returns a set of statistics that describes how often and for how long various parts of the program executed.
First, load python3 module:
$ module load python3
Then, use -m cProfile option of the python3 command:
$ python3 -m cProfile monte_carlo_pi.py
Sorting by cumulative:
Estimated value of pi: 3.1413716
40001353 function calls (40001326 primitive calls) in 17.496 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
3/1 0.000 0.000 17.496 17.496 {built-in method builtins.exec}
1 0.000 0.000 17.496 17.496 monte_carlo_pi.py:1(<module>)
1 0.000 0.000 17.489 17.489 monte_carlo_pi.py:16(main)
1 9.232 9.232 17.489 17.489 monte_carlo_pi.py:3(estimate_pi)
20000000 6.687 0.000 8.257 0.000 random.py:415(uniform)
The output can be ordered by several keys, use the -s option to select:
$ python3 -m cProfile -s cumulative monte_carlo_pi.py
Here is a table of acceptable keys:
Sort Key |
Description |
|---|---|
calls |
Number of times the function was called |
cumulative |
Total time spent in the function and all the functions it calls |
cumtime |
Total time spent in the function and all the functions it calls |
file |
Filename where the function is defined |
filename |
Filename where the function is defined |
module |
Filename where the function is defined |
ncalls |
Number of times the function was called |
pcalls |
Number of primitive (i.e., not induced via recursion) function calls |
line |
Line number in the file where the function is defined |
name |
Name of the function |
nfl |
Combination of function name, filename, and line number |
stdname |
Standard function name (a more readable version of ‘nfl’) |
time |
Internal time spent in the function excluding time made in calls to sub-functions |
tottime |
Internal time spent in the function excluding time made in calls to sub-functions |
Functions profiling
cProfile is a powerful tool for profiling Python scripts. However, it’s not always efficient to profile an entire script, especially if you have a specific function suspected to be a performance bottleneck.
In computational programs, a heavy-lifting function, such as one performing complex calculations or data manipulations, often consumes a significant portion of the execution time. Profiling this specific function can provide more granular insights, assisting in identifying areas for optimization.
Here’s how to use cProfile for a specific function:
import cProfile
def my_compute_func():
# Complex computation function
cProfile.run('my_compute_func()', sort='cumulative')
Targeted profiling helps optimize the most time-consuming parts of your code, potentially resulting in substantial improvements in the overall execution time.
cProfile and mpi4py
It is possible to profile MPI code using cProfile, but it can be a bit more complex than profiling a single process Python script.
When an MPI program is run, multiple processes are created, each with its own Python interpreter. If you just run cProfile as usual, each process will attempt to write its profiling data to the same file, which will lead to problems.
To avoid this, you can modify your MPI script to write the profiling data from each process to a different file. Here is an example using mpi4py:
from mpi4py import MPI
import cProfile
def main(rank):
# Your MPI code here
pass
if __name__ == "__main__":
comm = MPI.COMM_WORLD
rank = comm.Get_rank()
cProfile.run('main(rank)', f'output_{rank}.pstats')
Please remember that this way, the result is a number of different files each representing the profiling results of one MPI process. They won’t give you a total view of the time spent across all processes, but you will be able to see what each individual process spent its time on.
cProfile and gprof2dot
cProfile output can be formatted into reports via the pstats module:
$ python3 -m cProfile -o output.pstats monte_carlo_pi.py
The pstats file is readable by gprof2dot:
$ module load gprof2dot
$ gprof2dot.py -f pstats output.pstats -o output.dot
$ dot -Tpng output.dot -o output.png
Then, display output.png with eog or display command.
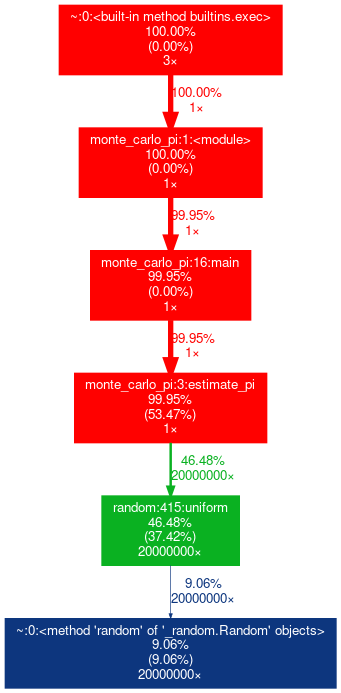
Nsight System
nsys is a profiler and graphical profile data analysis tool for CUDA applications.
It is installed with the Nvidia HPC Software Development Kit and can be accessed through the nvhpc module.
In order to profile your application, you can use the following startup command in a job script :
ccc_mprun nsys profile --trace=openacc,cuda,nvtx --stats=true ./set_visible_device.sh ./myprogram`
Warning
A known bug affects nsys MPI auto-detection feature when using OpenMPI 4.1.x versions which produces a segmentation fault.
In order to circumvent it, use the --mpi-impl=openmpi option when using --trace=mpi.
During execution, nsys will creates report<n>.sqlite and report<n>.nsys-rep files.
You may then use the command line or graphical interface to analyse and display these results.
nsys-ui starts up the graphical interface.
Use nsys-ui path/to/report*.nsys-rep to open up the nsys report files and navigate the collected traces.
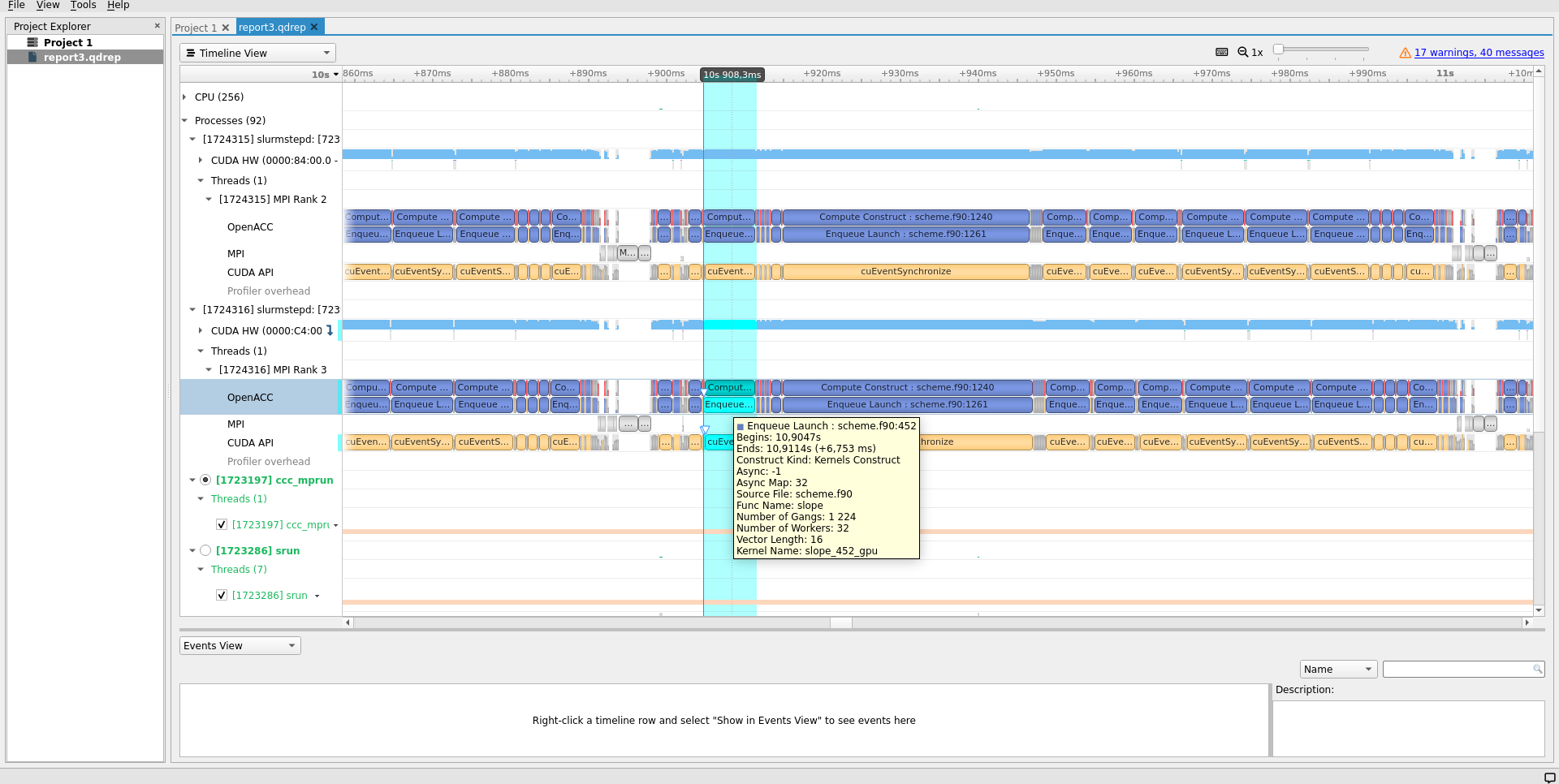
For convenience, you may install Nsight Systems on your own computer and copy the instrumentation data there for displaying.
Nsight Systems is freely available to members of the NVIDIA Developer Program which is a free registration.
Note that you will need to select the right version for download.
Use nsys --version to know which one is being used in your profiling jobs.
The nsys-ui graphical interface can also be used to launch computations directly on nodes reserved, eg. using ccc_mprun -K.
Refer to the online Nsight Systems documentation or to the nsys --help command for more information
Warning
When using nsys, the /tmp directory might become saturated, leading to process failures. To avoid this, it is recommended to redirect temporary files to another location by modifying the TMPDIR environment variable.
For example:
export TMPDIR=$CCCSCRATCHDIR
Nsight Compute
ncu is a cuda kernel profiler for analysing GPU occupancy, and the usage of memory and compute units. It features advanced performance analysis tools and a graphical reporting interface providing useful hints and graphs.
In order to profile your application kernels, you may use the following startup command in a job script :
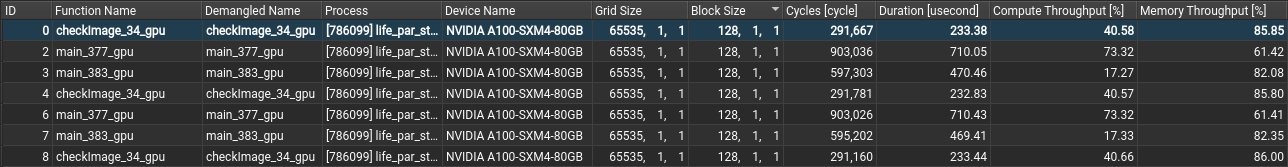
ccc_mprun ncu --call-stack --nvtx --set full -s 20 -c 20 --target-processes all -o report_%q{SLURM_PROCID} ./set_visible_device.sh ./myprogram
This example will generate one report file, named report_X.ncu-rep, for each process started by the application.
It uses the full argument to the --set option, which triggers the collection of a set of metrics.
You may use other values for the --set option for specifying a chosen set of sections, use --section for enabling individual report sections and the associated metrics, or use --metric for activating the collection of individual metrics.
Use ncu --list-sets, ncu --list-sections and ncu --list-metrics respectively for more informations.
The option -s 20 ignores the first 20 kernels executed, in an attempt to avoid gathering information on kernels which are not representative of the general workload, such as those in charge of data initialization.
The option -c 20 enables the profiling and collection of selected performance metrics for the next 20 kernels, providing control over the general overhead of profiling the executed program.
For more information, run ncu --help or refer to the online Nsight Compute CLI documentation.
Use the ncu-ui command to open the generated reports in the Nsight Compute GUI, and access the various performance analysis, hints, and graphs.
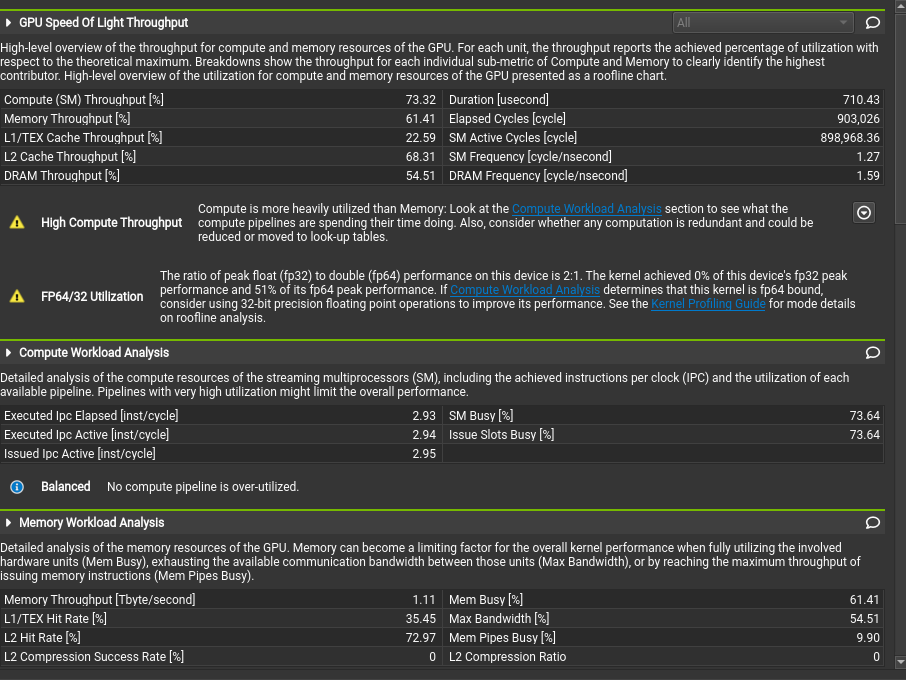
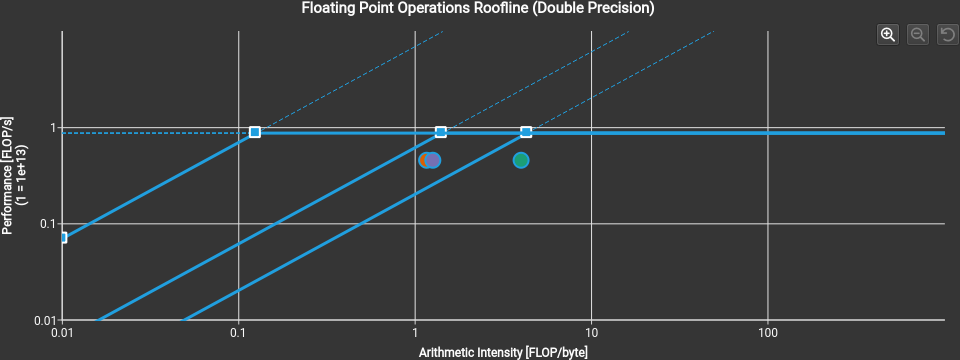
For more information, please refer to the online Nsight Compute documentation